Here is a sampling of my projects, past and present.
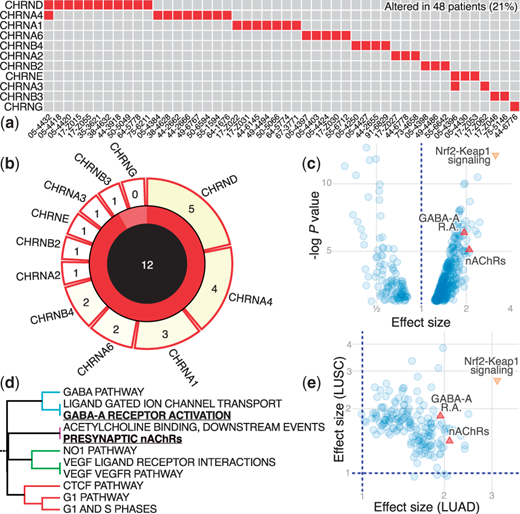
PathScore: a pathway analysis web app for cancer mutation datasets
PathScore calculates mutation enrichment across a database of curated pathways (MSigDB), controlling for background mutation rate, patient mutational load, and gene length. Interactive visualizations (powered by Bokeh) allow users to browse and search pathways, view mutations and compare projects. I designed the algorithm with Jeffrey Townsend, and built the web application using Flask and MySQL. Paper in Bioinformatics.
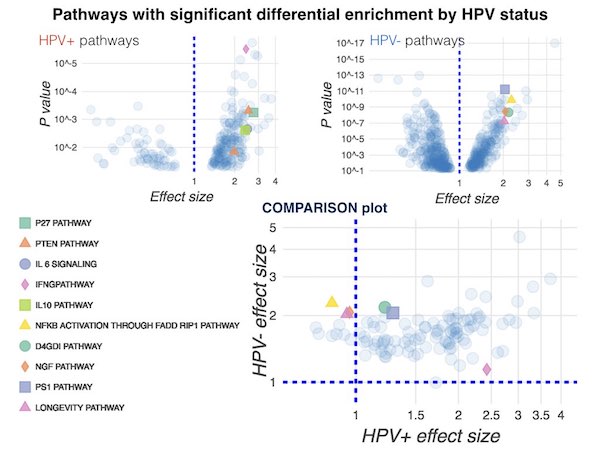
Cancer subtype comparisons
A recurring theme in my cancer projects is the identification of patterns that distinguish between patient subgroups. Such subgroups comparisons include: responders and non-responders to therapy; radiation-induced vs non-radiation-induced meningiomas; melanomas with and without key driver mutations, and lung cancer brain metastases and primary tumors.
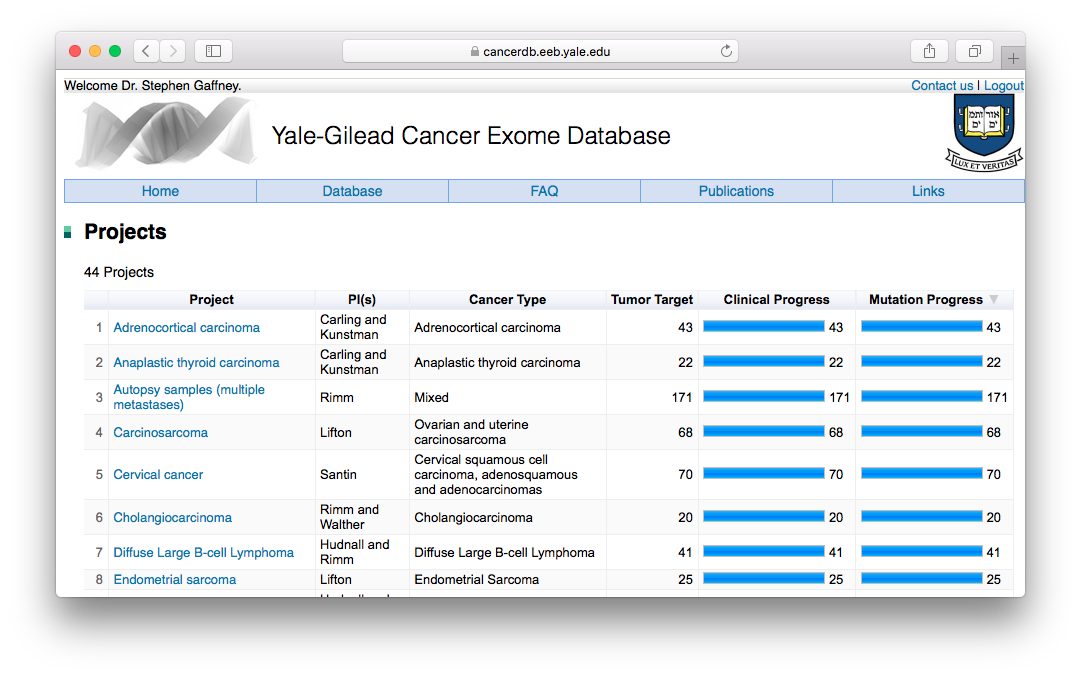
YCGED: Yale-Gilead Cancer Exome Database
A secure database for archiving and sharing somatic and germline mutations and corresponding clinical data from the Yale-Gilead collaboration. I designed the MySQL database and built the web application in Java Server Pages.
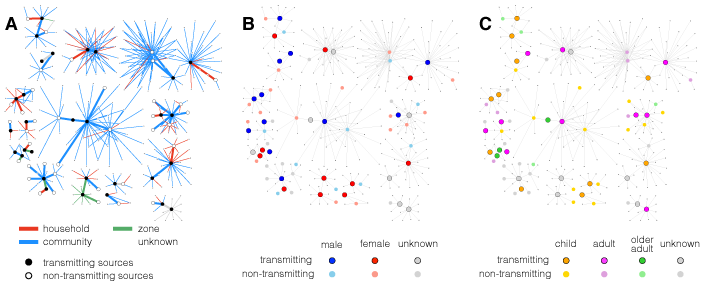
Network visualisation of Ebola contact tracing data
I contributed Ebola contact network plots and data analysis for an article in Philosophical Transactions B.
Skrip L, Fallah M, Gaffney SG, Yaari R, Yamin D, Huppert A, Galvani AP (2017) Characterizing risk of Ebola transmission based on frequency and type of case-contact exposures. Philosophical Transactions of the Royal Society B 372 (1721), 20160301
GEM-NET
I served as lead software engineer for the NSF Center for Genetically Encoded Materials (C-GEM), a collaboration between Yale, Berkeley and Stanford that is developing technology to programmatically build sequence-defined chemical polymers.
My role was to design their collaboration software infrastructure, build their website, and build data management systems to track their expanding biospecimen and chemical libraries. I documented this process and the tools involved in an article in ACS Central Science (Gaffney et al, 2019).
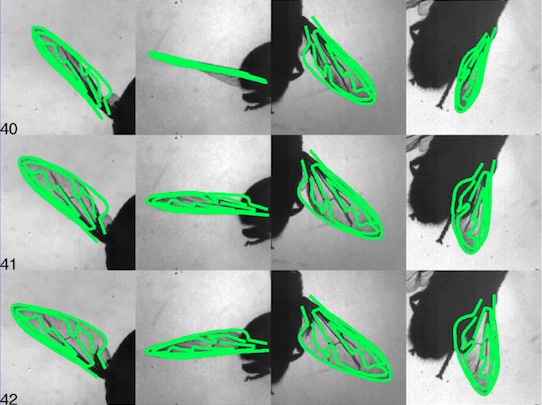
PhD: Markerless deformation capture of hoverfly wings using multiple calibrated cameras
In my PhD, I developed an algorithm for tracking the flapping flight of hoverfly wings in 3D, from high speed video. You can read my thesis here, and see the 3D shape from one of these images by clicking below.